2.40. MedeA : Polymer Expert De Novo Polymer Design
Contents
| download: | pdf |
|---|
2.40.1. Key Benefits of MedeA Polymer Expert
- Access to an extensive and carefully curated library of polymer repeat units.
- An analog targeted database with more than 1.1 million entries spanning diverse chemical variants based on this library.
- A flexible interface to construct queries based on similarity and adherence to defined property criteria.
- An efficient search engine searching millions of repeat units in property space to identify relevant entries.
- Access to functionality in both interactive user interface and Flowchart based modes.
- Full integration with MedeA structure lists, user interface, polymer model construction, and P3C functionality .
- The ability to identify and design polymers with desired properties.
- The ability to identify and design polymers derived from biobased starting points.
2.40.2. Computational Characteristics
- MedeA Polymer Expert employs a database of 1.1 million repeat units with each entry possessing information about its topology and chemistry structure, whether it was derived from a biobased starting point, and P3C based properties.
- The database can be searched on the basis of properties by similarity and mathematical conditions, such as less than or greater than desired values.
- Searches are performed rapidly and results supplied to the user in the form of structure lists .
2.40.3. Introduction
Despite considerable experimental and computational effort, the full design space of synthesizable polymeric systems has been explored to only a very limited extent. The need to produce high value polymers with targeted properties, to reduce reliance on precursors derived from fossil fuel stocks, and to create biodegradable plastics has led to a renewed interest in the design and development of novel polymeric systems.
2.40.4. Implementation
Using Materials Informatics methodologies \(^{1-15}\) MedeA Polymer Expert provides a unique methodology for the development of polymer systems with desirable property profiles. The implementation of MedeA Polymer Expert can be summarized as follows:
An initial database has been assembled which captures the chemical diversity of the polymer design space spanning the wide range of reported and characterized repeat units in the polymer literature.
A large analog database has been constructed starting from this initial literature work through the use of fragment additions.
Following construction of a 1.1 million repeat library, the P3C methodology has been employed to annotate each of these structures with QSPR-derived properties. The resulting library, PEARL (Polymer Expert Analog Repeat unit Library, is a rich source of chemical property information for use in the design and analysis of polymeric systems.
MedeA Polymer Expert possesses two means of addressing the PEARL database. These modes allow:
- Identification of repeat units possessing desired properties through the use of specified poperty targets, and the,
- Identification of polymers through the specification of target analogs
MedeA Polymer Expert queries can combine both modes of addressing the PEARL database.
MedeA Polymer Expert was developed in collaboration with Dr. Jozef Bicerano and makes extensive use of the expertise and correlations described in Prediction of Polymer Properties [16]. Additional information on the development, implementation, and application of MedeA Polymer Expert is available in the peer reviewed literature [17].
2.40.5. Analog Library
The basis for the construction of PEARL is shown in the figure below. A defined set of repeat units taken from the current research literature was systematically combined with commonly found organic fragments and groups. Full details of the construction of PEARL are provided in a paper describing the development of the methodology [17].

A schematic showing the basis for the construction of PEARL starting from a “seed” repeat unit and replacing a randomly selected hydrogen with a fragment from a fragment library in successive steps. Here the repeat unit of poly(isosorbyl carbonate) (PIC) is Generation 0 and is taken directly from the initial database. A repeat unit generated after 1, 2, or 3 iterative steps starting from the PIC repeat unit is labeled as a Generation 1, 2, or 3 derivatives of PIC. Only Generation 0, 1, and 2 repeat units were included in the PEARL final database to maximize chemical property space coverage without introducing undue synthetic complexity [17].
Note
MedeA Polymer Expert is the name of the MedeA module providing query building, database searching, and reporting capabilities for de Novo polymer design. PEARL (Polymer Expert Analog Repeat unit Library) is the name of the database employed by MedeA Polymer Expert.
2.40.6. Usage
Invoke MedeA Polymer Expert either interactively using the Polymer Expert entry in the MedeA/Tools menu, or automatically within the MedeA Flowchart interface.
- Interactively:
- Use the MedeA Tools menu to invoke MedeA Polymer Expert. by clicking on the appropriate entry. A query build and results view panel as shown below will be created in your MedeA environment.

- In a MedeA Flowchart:
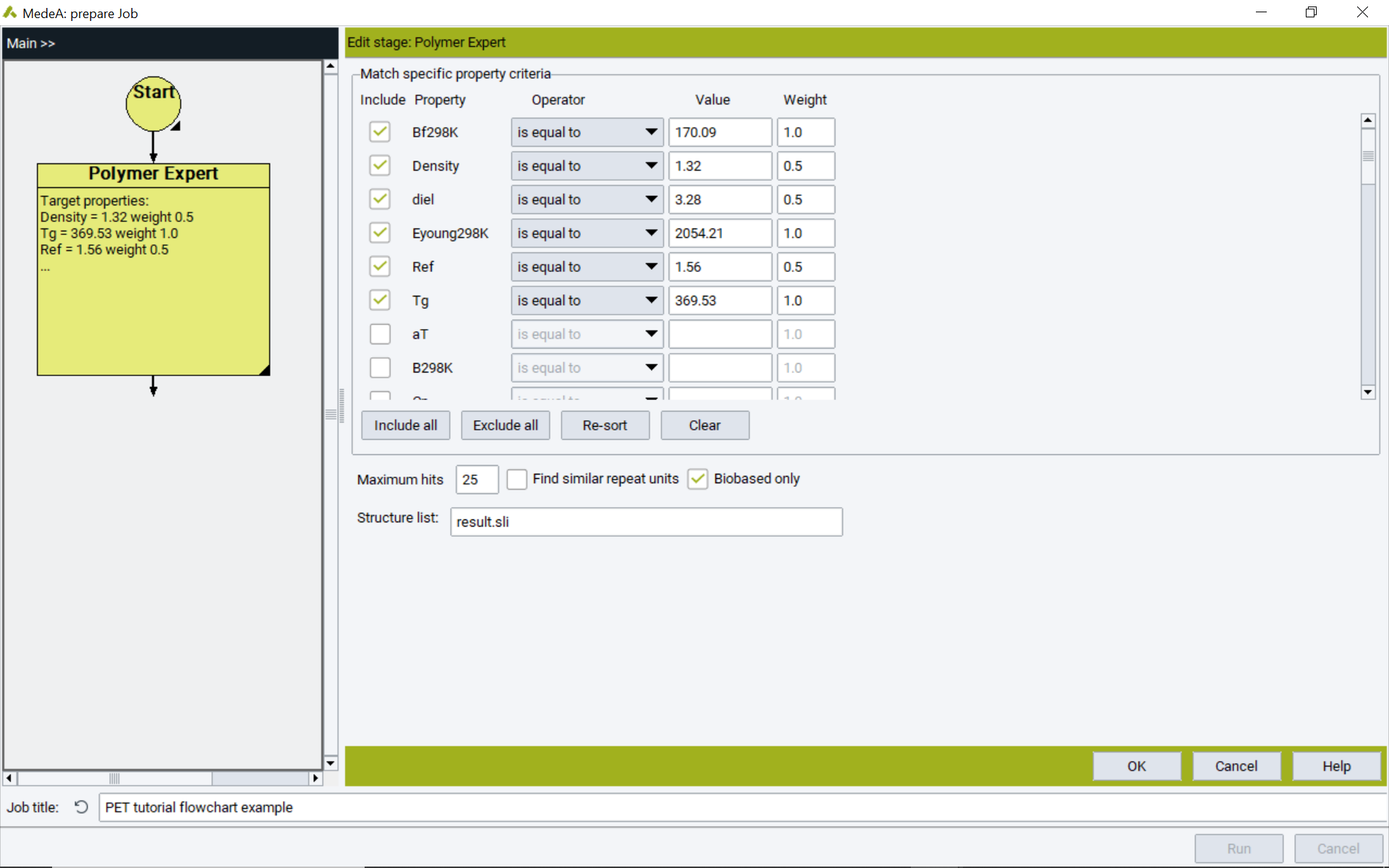
- Open the MedeA Flowchart via Job >> New Job…
- Add a MedeA Polymer Expert stage to the Flowchart, specify the search criteria, and click Run
- Upon completion of the Job, results are saved in the Job.out file
Note
One advantage of using MedeA Polymer Expert in a flowchart is that you can store query parameters as a permanent record of your query on the Job Server as a part of running a Polymer Expert stage. You can restore a given query using the Flowchart interface.
Note
One advantage of using MedeA Polymer Expert in its interactive form is that you can rapidly assess the affect of query details on the result set interactively. Polymer Expert queries on the PEARL database are typically completed in significantly less than one second and the result set can be investigated interactively in the MedeA interface.
2.40.7. Background
As described above, MedeA Polymer Expert searches the PEARL database in one of two ways. In similarity search mode, a similarity index, \(R^{i}\), is computed for each entry in the database using the equation below:
Here \(R^{2}\) is the similarity score or index, and \(w_{i}\), \(P_{ic}\), and \(P_{it}\) denote, respectively, the weight and values of the \(i^{th}\) property, computed and stored in PEARL and specified as the target of the query. A similarity score of 0 indicates exact agreement between computed and target properties. Query results produced by MedeA Polymer Expert are provided in similarity sorted order with the best agreement with the target, i.e. the lowest score, being first in the result set.
MedeA Polymer Expert can also query PEARL using the logical operations ‘is between’, ‘is less then’, ‘is greater than’, and whether a given repeat unit is derived from a biobased starting point. These query modes do not employ a supplied weight and do not result in a modification to the \(R^{2}\) similarity score or index for a given search result, they are instead logical operations.
Note
Logical operations may be combined with similarity score operations in MedeA Polymer Expert queries. When combined queries are performed, the result set is filtered using \(R^{2}\) as is the case for an exclusively similarity based search. For combined queries, all reported results will additionally obey the supplied property constraints.
Note
When used in a flowchart MedeA Polymer Expert reports the total number of PEARL database members that satisfy the supplied logical query. However, only the supplied Maximum hits number of repeat units will be saved by the job.
2.40.8. Output
When employed in a flowchart, MedeA Polymer Expert output in Job.out begins with a listing of the query employed in the PEARL search. This is followed by a listing of the hits obtained in the search, and additional information related to the search, including the execution time for the search and the name of the structure list file employed in storing the results of the search.
| [1] | Materials genome initiative strategic plan, a report by the Subcommittee on the Materials Genome Initiative, Committee on Technology of the National Science and Technology Council (USA), November 2021. |
| [2] | A. Chandrasekaran, C. Kim, R. Ramprasad (2020). Polymer genome: a polymer informatics platform to accelerate polymer discovery. In: K. Schütt, S. Chmiela, O. von Lilienfeld, A. Tkatchenko, K. Tsuda, K. R. Müller (eds), Machine Learning Meets Quantum Physics. Lecture Notes in Physics, vol. 968. Springer. |
| [3] | H. D. Tran, C. Kim , L. Chen, A. Chandrasekaran, R. Batra, S. Venkatram, D. Kamal, J. P. Lightstone, R. Gurnani, P. Shetty, Machine-learning predictions of polymer properties with polymer genome, J. Appl. Phys. 128 (17) (2020) 171104. |
| [4] | M. M. Cencer, J. S. Moore, R. S. Assary, Machine learning for polymeric materials: an introduction, Polym. Int. 71 (5) (2022) 537–542. |
| [5] | Y. Liu, T. Zhao, W. Ju, S. Shi, Materials discovery and design using machine learning, J Materiomics 3 (2017) 159-177. |
| [6] | W. Sha, Y. Li, S. Tang, J. Tian, Y. Zhao, Y. Guo, W. Zhang, X. Zhang, S. Lu, Y.-C. Cao, S. Cheng, Machine learning in polymer informatics, InfoMat. (2021) 3:353–361. |
| [7] | S. Wu, Y. Kondo, M. Kakimoto, B. Yang, H. Yamada, I. Kuwajima, G. Lambard, K. Hongo, Y. Xu, J. Shiomi, C. Schick, J. Morikawa, R. Yoshida, Machine-learning-assisted discovery of polymers with high thermal conductivity using a molecular design algorithm, npj Computational Materials (2019) 5:66. |
| [8] | M. Ruimin, L. Tengfei, PI1M: a benchmark database for polymer informatics, J. Chem. Inf. Model. (2020) 60, 4684−4690. |
| [9] | N. Meftahi, M. Klymenko, A. J. Christofferson, U. Bach, D. A. Winkler, S. P. Russo, Machine learning property prediction for organic photovoltaic devices, npj Computational Materials (2020) 6:166. |
| [10] | L. Chen, G. Pilania, R. Batra, T. D. Huan, C. Kim, C. Kuenneth, R. Ramprasad, Polymer informatics: current status and critical next steps, Materials Science and Engineering: R: Reports (2021) 144, 100595. |
| [11] | R. Gurnani, C. Kuenneth, A. Toland, R. Ramprasad, Polymer informatics at scale with multitask graph neural networks, Chemistry of Materials (2023) 35, 4, 1560-1567. |
| [12] | S. Wu, H. Yamada, Y. Hayashi, M. Zamengo, R. Yoshida, Potentials and challenges of polymer informatics: exploiting machine learning for polymer design (2020) https://arxiv.org/pdf/2010.07683.pdf. This is an English translation of the Japanese manuscript published in Proceedings of the Institute of Statistical Mathematics (2021 special issue). |
| [13] | D. J. Audus, J. J. de Pablo, Polymer informatics: opportunities and challenges, ACS Macro Lett. (2017) 6, 1078-1082. |
| [14] | L. Tao, V. Varshney, Y. Li, Benchmarking machine learning models for polymer informatics: an example of glass transition temperature, J. Chem. Inf. Model. (2021) 61, 5395−5413. |
| [15] | K. Hatakeyama-Sato, Recent advances and challenges in experiment-oriented polymer informatics, Polymer Journal (2023) 55, 117–131. |
| [16] | Jozef Bicerano, Prediction of Polymer Properties, Third. (New York: Marcel Dekker, Inc., 2002). |
| [17] | (1, 2, 3) J. Bicerano, D. Rigby, C. Freeman, B. Leblanc, and J. Aubry, Polymer Expert - A Software Tool for De Novo Polymer Design, submitted, (2023). |
| download: | pdf |
|---|